21 November 2019 by Remco Bouckaert
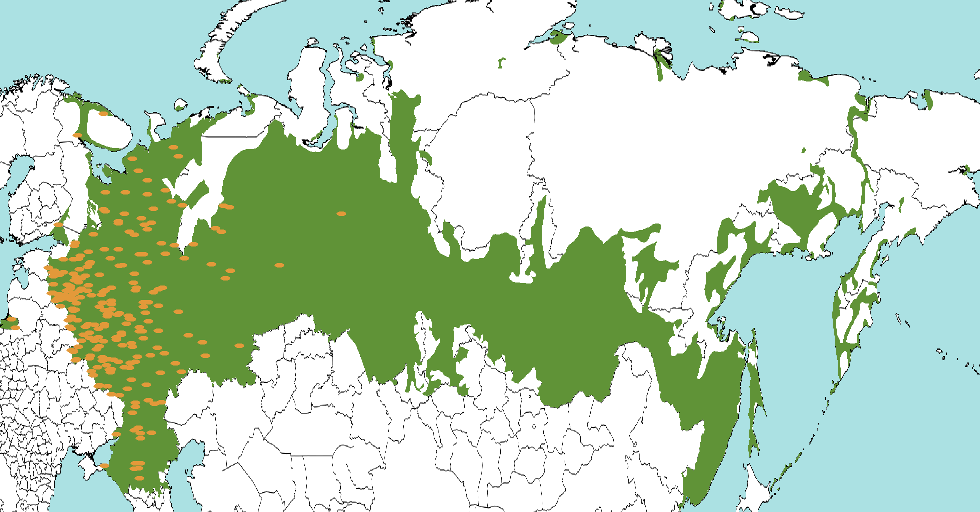
The GEO-SPHERE package can be used to perform phylogeography analysis assuming a random walk over a sphere – our planet. Here, we show how to define some ways you can define priors to restrict the location of some nodes in the tree. This allows for example tips to be located in a region that contains a species, instead of having to approximate it by a point location. For example, the green area below defines locations where Russian is spoken, and form the prior of a Russian taxon. Orange dots indicate posterior sample locations which are all to the western side. Note that locations do not need to be contiguous, and there are two samples in Kaliningrad next to the Baltic Sea.
Priors will be uniform for a given area as in Bouckaert et al, 2012, Nylinder et al, 2014, and do not allow heterogeneity within a region as in Dellicour et al, 2019.
Priors on tip locations
At this point in time, there is no BEAUti support to allow sampling of tip locations, but it can be done by editing the XML once it is produced by BEAUti.
First, you need to add an entry to the state, and add operators. Second, you have to define a set of regions, either by KML or by colouring a bitmap image in Mercatore projection. KML regions can be drawn in google-earth and bitmap images in Mercator projection can be produced using the MapMaker utility that you can start from the BEAST app launcher. Once you created a world map with MapMaker, you can colour in the region you want to sample from.
To set up the state, add the following line (where “geo” is the name used for the geography partition)
<stateNode idref="location.s:geo"/>
Note that the “s:" in “location.s:geo" is important! Next, add a
LocationOperator to the set of operators (inside the MCMC element, where
the other operators are):
<operator id='location.sampler' spec='sphericalGeo.LocationOperator'
location='@location.s:geo'
likelihood='@slocationtreeLikelihood.geo' weight='30'/>
Also, add a reference to location.s:geo in
ApproxMultivariateTraitLikelihood by adding the attribute
location=“@location.s:geo” to the element with
id=“slocationtreeLikelihood.geo”.
To specify a KML region, add the following fragment to the XML at the top level (just under the alignment, but above the run element would be a good place).
<region id='China' spec='sphericalGeo.region.KMLRegion' kml='kml/China.kml'/>
This creates a KMLRegion with id China, and reads in the kml file in the
folder kml from file China.kml. To create a bitmap region from a bitmap
file stored in say regions/China.png, you can use
<region id='China' spec='sphericalGeo.region.BitMapRegion'
file='regions/China.png'/>
There is an extra option to specify a bounding box by two latitude/longitude pairs if the bitmap only covers a small region of the map instead of the whole world:
<region id='China' spec='sphericalGeo.region.BitMapRegion'
file='regions/China.png' bbox=''/>
Now we defined a region, suppose you want to sample taxon AF182802 from the region of China, you add
<geoprior location='@location.s:geo' tree='@Tree.t:hbv' region='@China'
spec='sphericalGeo.GeoPrior'>
<taxon id='AF182802' spec='Taxon'/>
<geoprior>
inside the element with id=’slocationtreeLikelihood.geo’ (where ’geo’ is
the name of the partition for spherical geography, and ‘hbv’ the name of
the partition with the alignment).
If you have many regions and tips you want to sample, to prettify the XML and make it somewhat more readable, you can create these maps:
<map name="region">sphericalGeo.region.KMLRegion</map>
<map name="geoprior">sphericalGeo.GeoPrior</map>
and then you can leave out the spec attribute and instead of the above use
<region id='China' kml='kml/China.kml'/>
and
<geoprior location='@location.s:geo' tree='@Tree.t:hbv' region='@China'>
<taxon id='AF182802' spec='Taxon'/>
<geoprior>
Finally, in order to initialise the locations and let the likelihood know which locations are sampled, and which are approximated directly, we have to do two things:
- let the
ApproxMultivariateTraitLikelihoodknow where the location parameter is by adding the location-attribute, and - add an entry for every
geoprior.
After this, the XML for the geography likelihood should look something like this:
<distribution id="geography" spec="sphericalGeo.ApproxMultivariateTraitLikelihood" tree="@Tree.t:AFRICA">
<location idref="location.s:geo">
<geoprior idref="China"/>
<geoprior idref="Eurasia"/>
...
Prior on the root locations
Putting a prior on the root means we have to sample the root location, which essentially follows the recipe for sampling tips, but instead of specifying a single taxon, we specify a taxon set containing all taxa. If we want to restrict the root location to China, as specified above, this can be done for example like so:
<geoprior location='@location.s:geo' tree='@Tree.t:hbv' region='@China'>
<taxonset id='all.taxa' spec='TaxonSet' alignment='@hbv'/>
<geoprior>
Prior on a monophyletic clade
First, specify a monophyletic clade in the priors panel in BEAUti; hit the little ’+’ button at the bottom, and a dialog pops up where you can specify a taxon set. Once you have given the clade a name (say “myclade”) and close the dialog, in the priors panel a new entry appears for the clade. Select the ’monophyletic’ check box. Note that the clade must be monophyletic, otherwise the sampler does not work.
To specify the prior, add the following to the element with
id=“slocationtreeLikelihood.geo”
<geoprior location='@location.s:geo' tree='@Tree.t:hbv' region='@China'>
<taxonset idref='myclade'/>
<geoprior>
References:
Bouckaert, R., Lemey, P., Dunn, M., Greenhill, S.J., Alekseyenko, A.V., Drummond, A.J., Gray, R.D., Suchard, M.A. and Atkinson, Q.D., 2012. Mapping the origins and expansion of the Indo-European language family. Science, 337(6097), pp.957-960.
Dellicour, S., Lemey, P., Artois, J., Lam, T.T., Fusaro, A., Monne, I., Cattoli, G., Kuznetsov, D., Xenarios, I., Dauphin, G. and Kalpravidh, W., 2019. Incorporating heterogeneous sampling probabilities in continuous phylogeographic inference–application to H5N1 spread in the Mekong region. Bioinformatics.
Nylinder S, Lemey P, De Bruyn M, Suchard MA, Pfeil BE, Walsh N, Anderberg AA., 2014. On the biogeography of Centipeda: a species-tree diffusion approach. Systematic Biology 63: 178-191.
