January 2023 by Remco Bouckaert
The default mixture of operators produced by BEAUti has changed from v2.6 to v2.7 in order exploit the latest developments in operator design. In particular,
- all standard operators have been replaced with Bactrian versions (Yang & Rodríguez, 2013, Thawornwattana et al, 2018),
- an adaptable variance multivariate normal (AVMN) operator (Baele et al, 2017) is added that updates multiple real valued parameters at the same time, and
- adaptable operators (Douglas, et al, 2021) are added that measure performance of a set of Bactrian and AVNM operators and reweigh them based on how often they are accepted, how big the changes are and how much time posterior recalculation takes.
- BICEPS operators for tree stretching and flexing (Bouckaert, 2022) replace the standard tree scaler.
In this post, we will have a look at the effect of these operators combined by comparing BEAUti v2.6 XML with BEAUti v2.7 XML.
Set up of comparison
To confirm an analysis has converged, we load the trace file in Tracer and check all parameters have reached an ESS of at least 200. This implies that one set of operators A is more efficient than another B if that goal has been reached in less time by A than by B. When parameters have a much larger ESS than 200, the operators on these parameters can be down-weighted in order to obtain better efficiency, since more effort can be put in increasing the ESS of other parameters. With this in mind, we measure efficiency in units of ESS per second.
Since ESS tends to change between runs, even when running the same number of MCMC samples, we run each analysis 10 times and average ESS over the 10 runs.
| dataset | taxa | sites | tip samples |
|---|---|---|---|
| HCV | 63 | 411 | no |
| Shankarappa’s Patient 9 (P9) | 117 | 663 | yes |
| COVID | 190 | 9763, 9756, 9750, 583 | yes |
We consider the following variants on the datasets listed above:
- v2.6: BEAUti v2.6,for HCV and Shankarappa’s Patient 9 (P9) using GTR + estimated frequencies + 4 gamma, Bayesian skyline tree prior and strict clock, for for COVID using four partitions with HKY + estimated frequencies, Bayesian skyline and strict clock.
- v2.7a: BEAUti 2.7 with same model but with Bactrian operators with same weights.
- v2.7b: As sv2.7a with AVMN and adaptable operators with same weights.
- v2.7c: As sv2.7b with reduced weights on real parameter operators (like birth rate, frequencies, clock rate, etc).
The last version (v2.7c) is currently the default in BEAUti.
Results
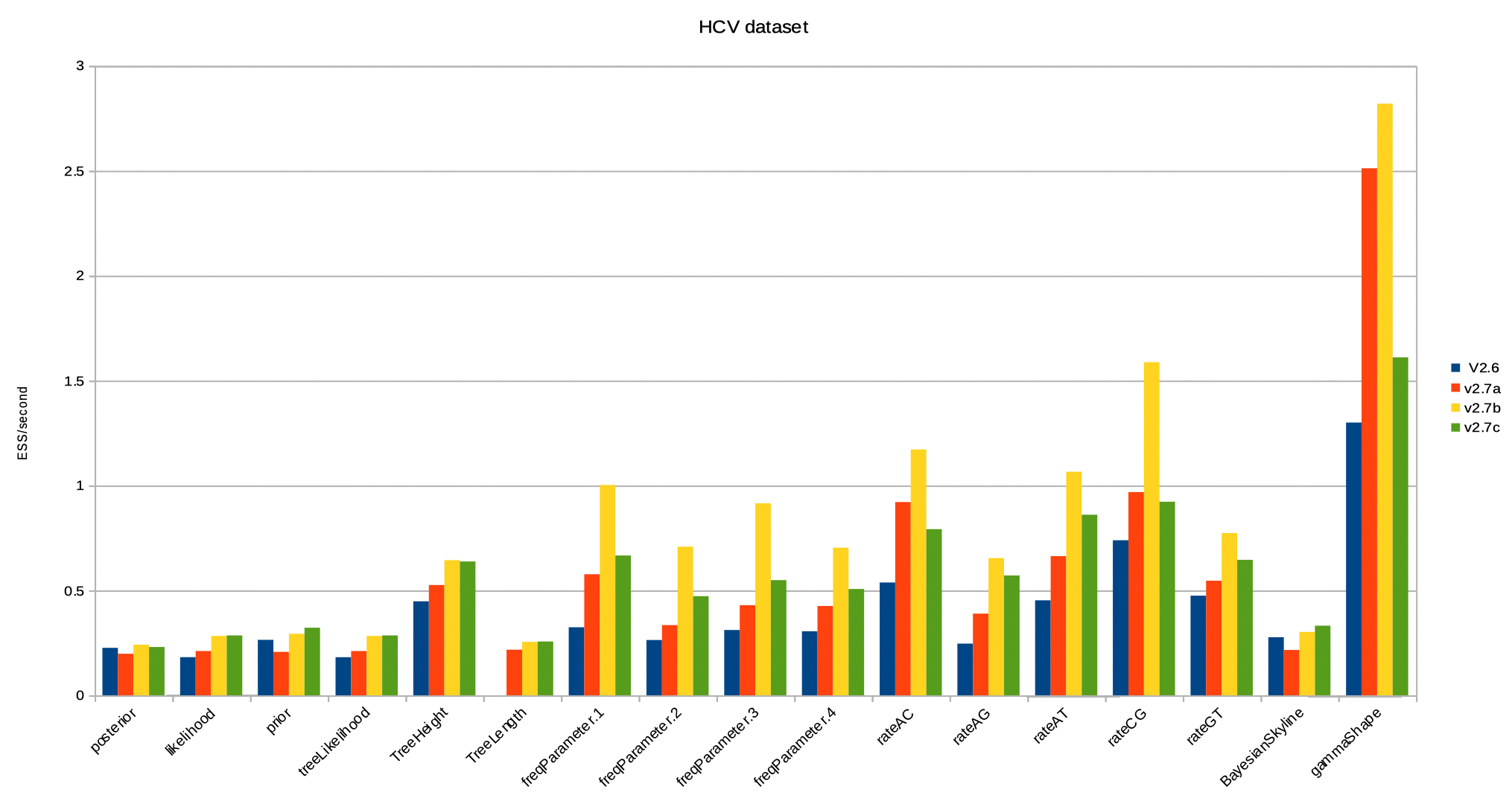
Below, the ESS per second for the HCV dataset. Some things to note:
- Bactrian operators alone (v2.7a vs v2.6) tend to increase ESS/second, with the exception of the tree prior.
- Adding the AVMN and adaptable operators (v2.7b vs v2.7a) increases the ESS/second even more for more real valued parameters, but not for the tree prior.
- reweighing (v2.7c vs v2.7b) gives overall a better balance of all items logged in the trace file.
- reweighing causes slow operators (like rate updates) to be replaced by faster operators (like subtree slide), thus slightly reducing over all runtime.
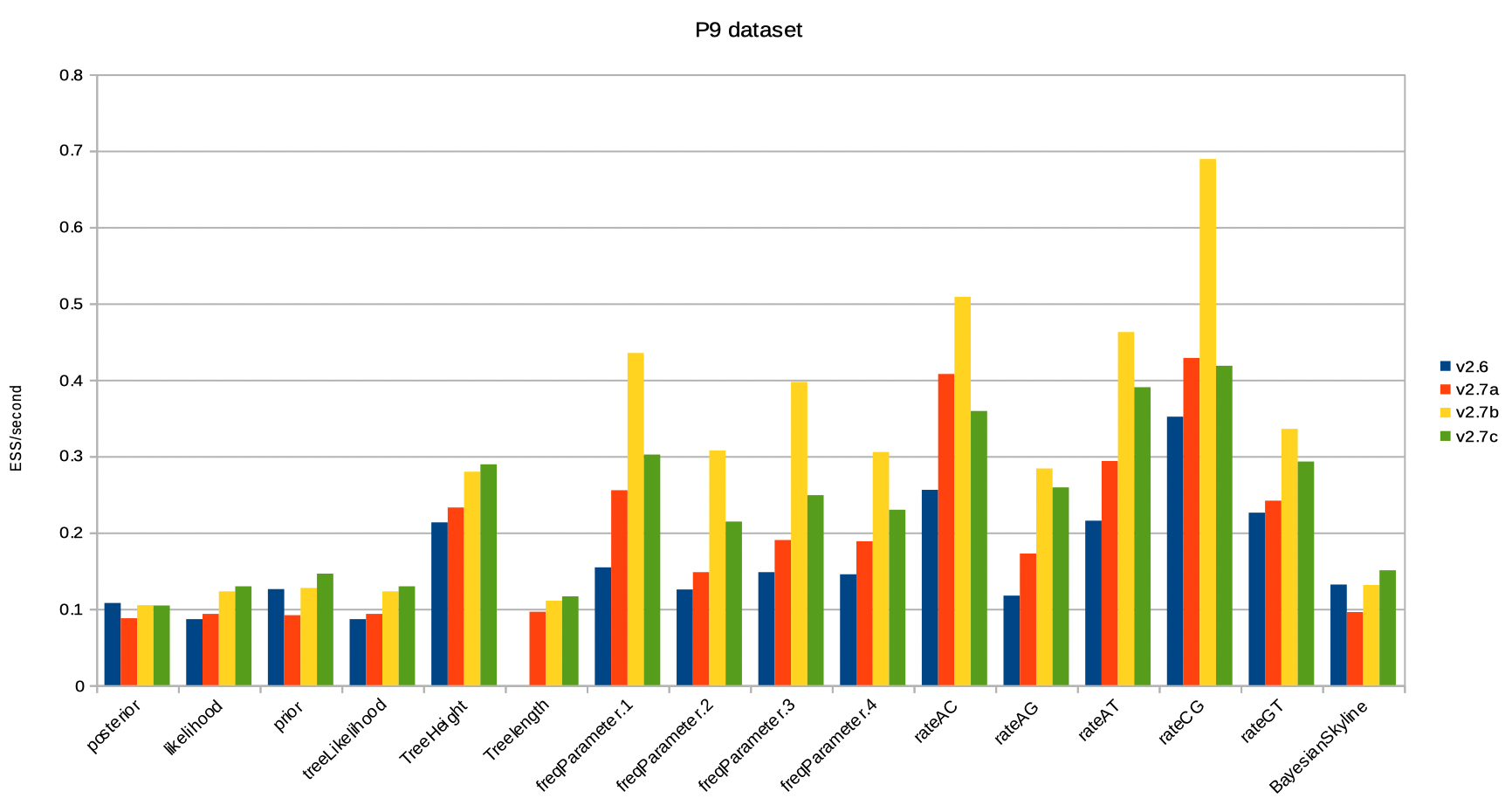
Results for the P9 dataset are very similar:
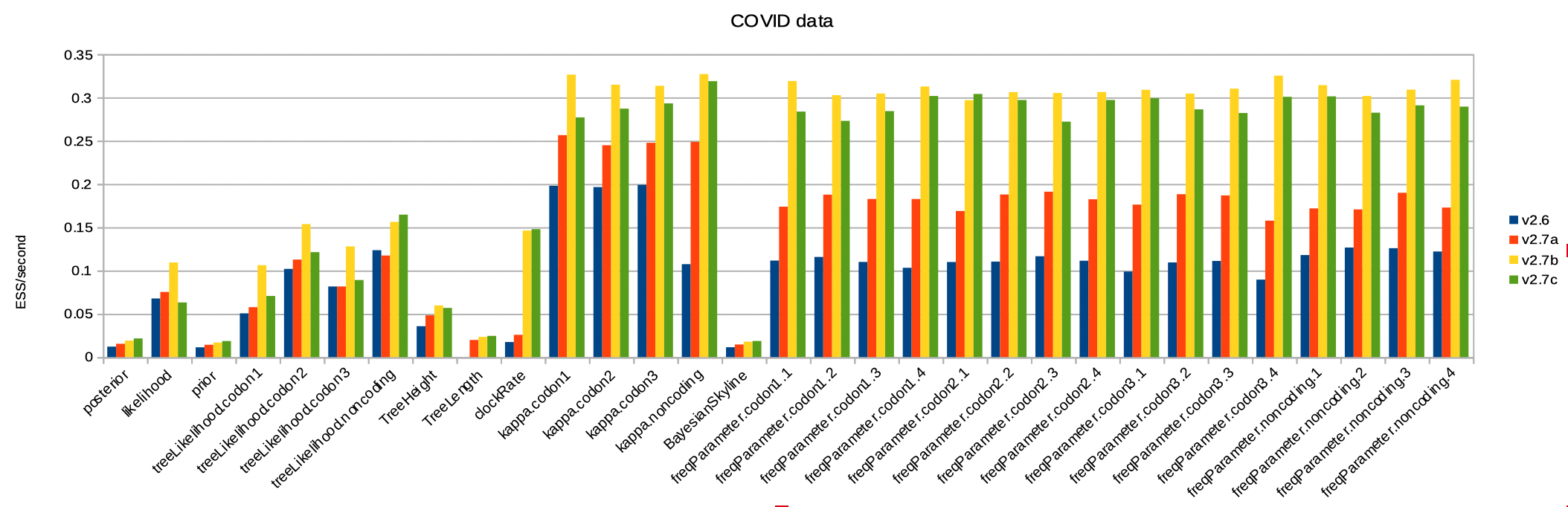
Here is the ESS per second for the COVID dataset. It benefits greatly from the BICEPS operators, but obviously benefits from the real valued parameter operators as well. Note here all ESSs per second increase, including the posterior and prior. Rates and frequencies increase much more, and arguably operators on there parameters can be down-weighted in order to increase ESS of prior and posterior even more.
Conclusions
- BEAST v2.7 seems to produce results faster than v2.6 for standard analyses.
- Package developers should consider using these new sets of operators where applicable.
- Tests were done with default operator weights as provided by BEAUti, but still can be changed and optimised by users.
XML files
Files used in this benchmark can be found here: https://github.com/CompEvol/benchmark/v2.6-vs-v2.7.
References
Baele G, Lemey P, Rambaut A, Suchard MA. Adaptive MCMC in Bayesian phylogenetics: an application to analyzing partitioned data in BEAST. Bioinformatics. 2017 Jun 15;33(12):1798-805. doi:10.1093/bioinformatics/btx088
Remco R Bouckaert. An Efficient Coalescent Epoch Model for Bayesian Phylogenetic Inference, Systematic Biology, syac015, 2022 DOI:10.1093/sysbio/syac015
Douglas J, Zhang R, Bouckaert R. Adaptive dating and fast proposals: Revisiting the phylogenetic relaxed clock model. PLoS computational biology. 2021 Feb 2;17(2):e1008322. doi: 10.1371/journal.pcbi.1008322
Thawornwattana Y, Dalquen D, Yang Z. Designing simple and efficient Markov chain Monte Carlo proposal kernels. Bayesian Analysis. 2018;13(4):1037-63. doi: 10.1214/17-BA1084
Yang Z, Rodríguez CE. Searching for efficient Markov chain Monte Carlo proposal kernels. Proceedings of the National Academy of Sciences. 2013 Nov 26;110(48):19307-12. doi: 10.1073/pnas.1311790110
