This post is in celebration of our paper on the origin and expansion of Pama-Nyungang languages, the largest language family of Australia that appeared today. In this paper, we introduced a new method of phylogeography: the founder-dispersal model aka the break-away model. A site that aims to make the material more accessible is here.
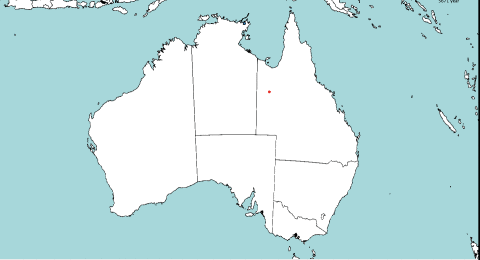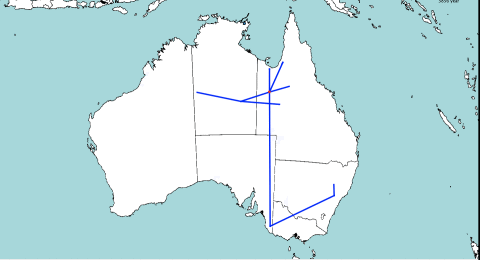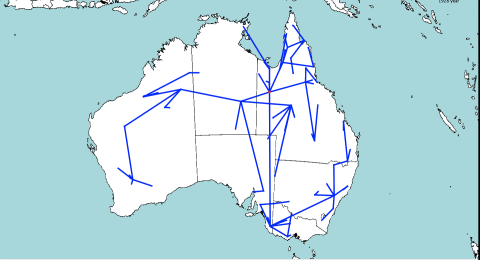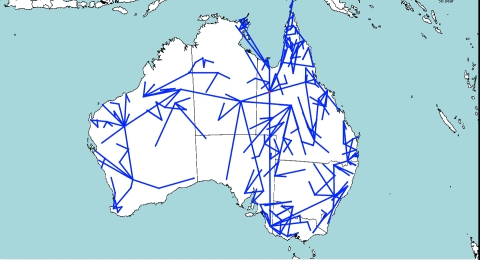
The idea behind the model is that at every split in the tree, one of the populations stays in the same place, and the other migrates. This is unlike other diffusion based methods, that assume that there is migration along both branches of the tree. It seems more intuitive that if a location contains a population, there must be a reason the population is there, e.g. a lot of resources being available. So, there should not be a reason for both populations to migrate, one of them could stay.

Known limitations
At the time of writing, there are a few limitations to this model.
Due to the lack of a good initialiser, this only works without tree initialisers, so you need to remove them from the XML. The tree does not need to be close to the posterior, it can be sampled from the prior, but the tree must not have a tree-initialiser.
Priors on the location of taxa or a clade (including root) are not implemented yet. This should not be hard once a good initialiser is developed.
Setting up an analysis
To set up an analysis, you need to install the break-away package. See here on how to install.
At the time of writing, there is no BEAUti support, so you have to edit the XML in order to do this. First, you can set up an analysis in BEAUti that contains all models and data that you want, except for the geography part. For branches that allow migration, we assume spherical diffusion, though the paper describes a few models that allow different rates depending on the underlying landscape -- but that will be for another post.
Step 1: remove tree initialisers
The stateNode containing the tree that will be used is typically initialised by aRandomTree inside an init element. Remove that element and if you have monophyletic or age constraints on the tree, use TreeParser in the tree stateNode. More about starting trees here.
Step 2: add location parameter
We need a location parameter, the precision for the diffusion model, and potentially a number of parameters for the clock model. Here, we add a strict clock, but a relaxed clock tends to fit well in general. For a strict clock, either the clock rate or the precision needs to be specified. Here, we specify the precision. Add the following two lines to the state:<stateNode id="location.BreakAway" spec='parameter.IntegerParameter' minordimension="1" dimension="8" value='0' estimate="true" />
<stateNode id="precision.s:BreakAway" spec="parameter.RealParameter" minordimension="1" estimate="true" value="0.001" lower="0.0"/>Step 3: add priors on parameters
We assume a uniform prior on the precision, with small lower bound and large upper bound to prevent numerical issues -- check that this is appropriate for your data by ensuring the distribution does not hug the upper or lower bound in Tracer. If so, these bounds may need changing.<prior id="precisionPrior.BreakAway" x="@precision.s:BreakAway" name="distribution">
<distr spec="Uniform" lower="0.001" upper="150000.0"/>
</prior>Step 4: add likelihood
We will assume that the tree partition is called tree. If your analysis uses a different name -- say DNA -- for the tree-partition, you need to change Tree.t:tree to Tree.t:DNA in the XML below. Furthermore, we assume there is a taxonset with id="TaxonSet.all" containing all taxa in the tree. You may need to change the reference to TaxonSet.all below if that taxonset has a different ID.
Locations are encoded as latitude-longitude pairs, and separated by a comma. Each of the taxa in the tree should have a location associated with it. Make sure to use correct spelling of taxon names.
<distribution id="geolikelihood" spec="beast.geo.SampledTraitLikelihood" tree="@Tree.t:tree">
<model id='model' spec='beast.geo.DistanceBasedDiffusionModel' precision="@precision.s:BreakAway" init='@location.BreakAway' tree='@Tree.t:tree' taxonset='@TaxonSet.all'>
<location spec="beast.geo.IslandHopLocationParameter" tree="@Tree.t:tree" location="@location.BreakAway"/>
Badimaya = -28.3 117.3,
ChampionBay = -28.8 114.5,
CentralAnmatyerr = -21.9 133.4,
<!-- more locations here, one for each taxon -->
</model>
<branchRateModel id="StrictClock.c:geo" spec="StrictClockModel" clock.rate="1.0"/>
</distribution>
Step 5: add operators
We add a scale operator for the precision, anIslandHopOperator that changes which branch stays and which one goes. Further, for every operator that changes the tree topology (like the Wilson Balding, wide and narrow exchange and subtree-slide operators) we add a IslandHopeMetaOperator, which changes the assignment of locations for only those nodes for which parents or children changed.
<operator id="precisionScaler.s:BreakAway" spec="ScaleOperator" parameter="@precision.s:BreakAway" weight="3" scaleFactor="0.7"/>
<operator id="islandHopper.s:BreakAway" spec="beast.geo.IslandHopOperator" tree="@Tree.t:tree" location="@location.BreakAway" weight="25"/>
<operator id="islandHopper.s:BreakAway" spec="beast.geo.IslandHopOperator" tree="@Tree.t:tree" location="@location.BreakAway" weight="25"/>
<!-- for every operator that change the tree topology
wrap it inside a IslandHopeMetaOperator
-->
<operator id="iHopSubtreeSlide" spec="beast.geo.IslandHopeMetaOperator" tree='@Tree.t:tree' location="@location.BreakAway" weight="15.0">
<operator id="SubtreeSlide.t:tree" spec="SubtreeSlide" tree="@Tree.t:tree" weight="15.0"/>
</operator>Step 6: add tree logger
Replace the tree logger with the tree logger shown below:<logger fileName="break-away.trees" id="treelog.t:BreakAway" logEvery="100000" mode="tree">
<log id="TreeWithMetaDataLogger.t:BreakAway" spec="beast.evolution.tree.TreeWithMetaDataLogger" tree="@Tree.t:tree" >
<metadata spec='beast.geo.LocationToLatLong2' id="location" model="@model" likelihood="@geolikelihood" value="0.0"/>
</log>
</logger>Analysing results
Since locations will be logged in the tree, just like for phylogeography based on diffusion on a plane or on a sphere, analysing the results is the same as for these other kind of phylogeography analyses. For example, the tutorial for diffusion on a sphere has more details.
