15 December 2020 by Jordan Douglas
The (uncorrelated) relaxed clock model is widely used in phylogenetics and enables the estimation of substitution rates across lineages (Drummond et al, 2006). It works well with when branch lengths and branch rates are not highly correlated (typical when sequences are not too large), so the fast relaxed clock model was developed (Zhang & Drummond) to remedy this. However, we found this implementation had performance issues where the previous model performed well.
ORC is a BEAST2 package which contains multiple performance optimisations related to this model (Douglas et al, 2020) including use of Bactrian proposals (Yang & Rodríguez, 2013). The operators included in ORC are especially efficient on long alignments, but even on short alignments perform better than the default relaxed clock model. Our operator scheme is adaptive and reacts to the dataset, ensuring that operators which bring about larger changes to the relaxed clock, in shorter computational runtime, are sampled with greater probabilities.
Setting up ORC
1) Launch BEAUti
2) To install ORC, click on File -> Manage Packages and install ORC.

3) Import an alignment as per usual (using File -> Import alignment)
4) On the Clock Model tab, select Optimised Relaxed Clock from the dropdown menu
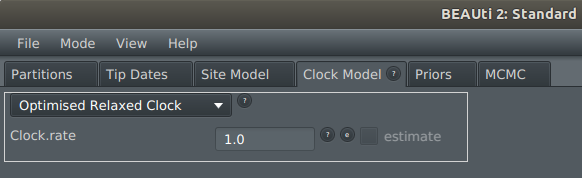
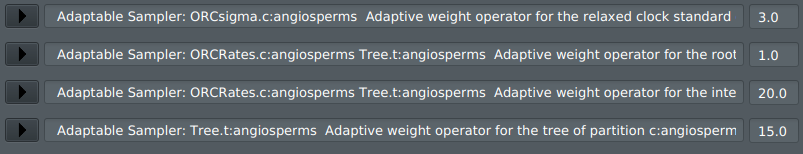
5) This should be working now. To confirm it has worked, display the Operators tab:

You should see the following 4 adaptive operators in the Operators tab:
6) Press File -> Save As and run the XML file as per usual
Parameters
Under the relaxed clock model employed by ORC, there is one substitution rate associated with each branch. These rates are distributed independently under a Log-Normal(mean=1, sigma) prior distribution for clock standard deviation sigma. Therefore 2N-2 rates (for taxon count N) and clock standard deviation sigma must be estimated.
Performance
Averaged across 8 biological datasets and 1 simulated dataset, ORC yielded a relaxed clock mixing rate between 1.2 and 13 times as fast as the standard BEAUti operator scheme. The improvement is stronger on long alignments. In the most extreme case we considered (a 7kb nucleotide alignment), the relaxed clock mixed up to 66 times faster under ORC, depending on the parameter. For further details, please see the Douglas et al. 2020.
Useful links
References
Douglas, Jordan, Rong Zhang, and Remco Bouckaert. Adaptive dating and fast proposals: Revisiting the phylogenetic relaxed clock model. PLoS computational biology. 2021 Feb 2;17(2):e1008322. DOI
Drummond, Alexei J., et al. “Relaxed phylogenetics and dating with confidence.” PLoS Biol 4.5 (2006): e88. DOI
Yang, Ziheng, and Carlos E. Rodríguez. “Searching for efficient Markov chain Monte Carlo proposal kernels.” Proceedings of the National Academy of Sciences 110.48 (2013): 19307-19312. DOI
Zhang, Rong, and Alexei Drummond. “Improving the performance of Bayesian phylogenetic inference under relaxed clock models.” BMC Evolutionary Biology 20 (2020): 1-28. DOI
