20 April 2020 by Remco Bouckaert
It is recommended to run multiple MCMC chains as a way to get confidence the MCMC algorithm converges on the same distribution. For trace logs, this is easily done by comparing distributions in Tracer. So, what to do with the tree sets? Here, we explain how to create plots like this and to see how much two tree distributions differ.
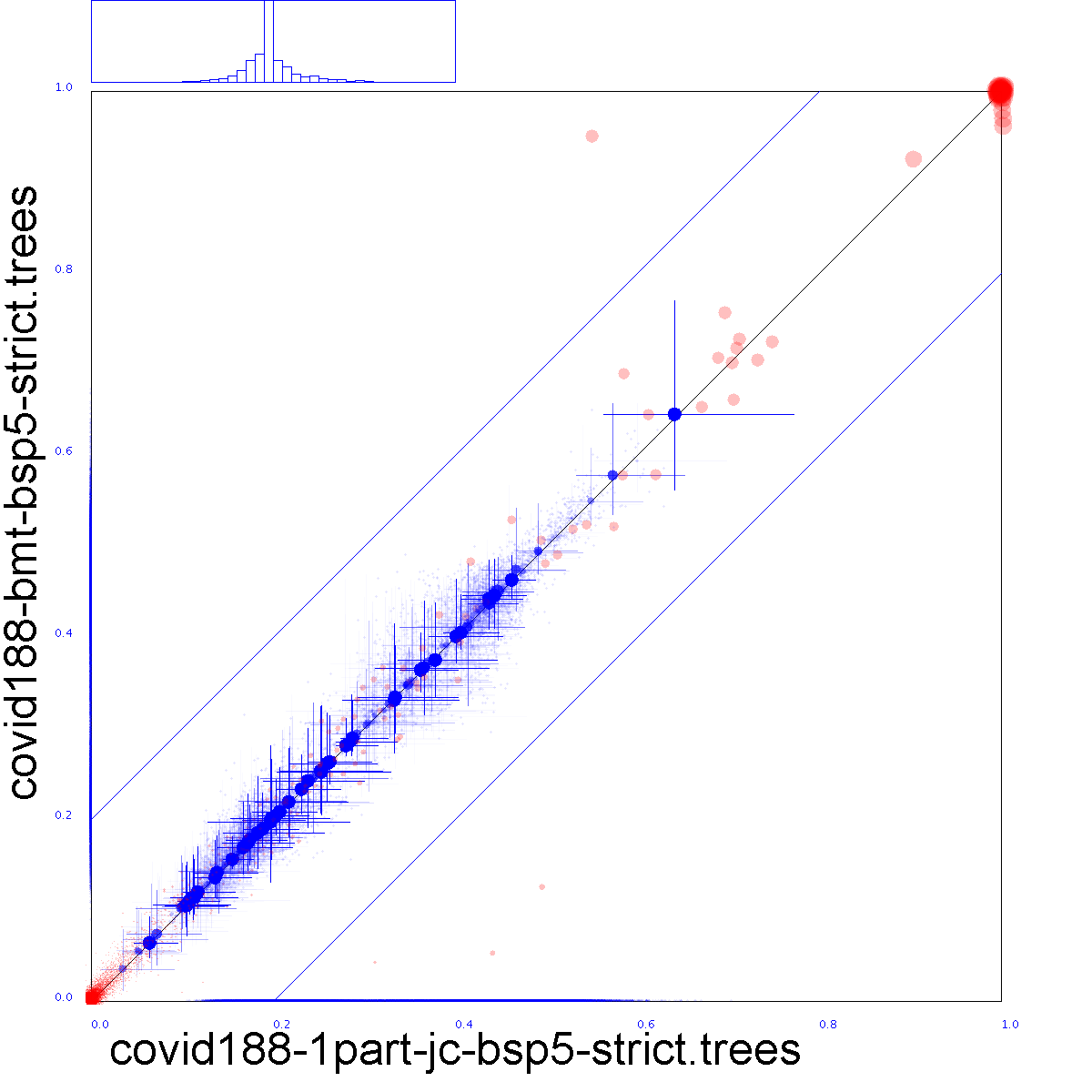
Red dots indicate clade support, running from zero probability (bottom left corner) to 1 (top right corner).
Blue dots indicate the mean of the height of a clade. Both x and y axis are scaled so that the largest tree height (from among both data sets) is at the top right corner. Blue crosses indicate 95% confidence interval of clade height.
For both red and blue dots, bigger dots mean more clade support.
The x-axis represent clade values from one set, the y-axis that of another. If both sets are the same, all dots will be on the x=y line (black diagonal line). The two blue lines indicate there is 20% difference: points within these lines have less than 20% difference, points outside have more, and should be considered as evidence there is substantial difference between the two posteriors.
The blue histogram at the top show the distribution of tree interval lengths for clades. Say, the interval size for a clade is of length A in the first set and B in the second, then if A>B, the first half of the histogram counts how often B/A is observed. However, if A<B then the second half of the graph counts how often A/B is observed. A more peaked histogram indicates less differences between tree interval sizes. If the graph is not symmetric but skewed to one side, it indicates one tree set has more uncertainty in interval estimates than the other.
Run CladeSetComparator
CladeSetComparator is a tool in Babel v0.3.0
(use the package manager
to install, or build Babel from source, and install by hand). Once it is installed, you
can run it via the File/Launch Apps menu in BEAUti, which then shows a dialog like so:
or on a terminal with the applauncer, e.g. like so:
/path/to/beast/bin/applauncher CladeSetComparator -tree1 tree1.trees -tree2 tree2.trees -png tree1vs2.png
where /path/to/beast the path to where BEAST is installed on your computer.
CladeSetComparator has the following options:
- -tree (TreeFile): 2 or more source tree (set or MCC tree) files (optional, but either -tree or -tree1 and -tree2 need to be specified)
- -tree1 (TreeFile): source tree (set or MCC tree) file (optional, but see -tree)
- -tree2 (TreeFile): source tree (set or MCC tree) file (optional, but see -tree)
- -out (OutFile): output file, or stdout if not specified (optional, default: [[none]])
- -svg (OutFile): svg output file. if not specified, no SVG output is produced. (optional, default: [[none]])
- -png (OutFile): png output file. if not specified, no PNG output is produced. (optional, default: [[none]])
- -burnin (Integer): percentage of trees to used as burn-in (and will be ignored) (optional, default: 10)
If you have the BEASTLabs (built from source or version >= 1.9.3) installed, you can process more than two tree sets like so:
/path/to/beast/bin/applauncher CladeSetComparator -tree tree1.trees tree2.trees tree3.trees -out out.txt -png tree.png
It will produce pair-wise comparisons for all tree sets and outputs
tree0-1.png, tree0-2.png and tree1-2.png as image files – note that tree.png
was specified as output file, but since there is more than one output when there are
more than 2 tree sets, the base-file name has tree set numbers added.
Also, a file tree.html will be produced that puts all png images in a table, for ease
of navigation, so it may look something like this:
Further out0-1.txt, out0-2.txt and out1-2.txt will be produced as text files with clade information.
| 0 | 1 |
|---|---|
 |
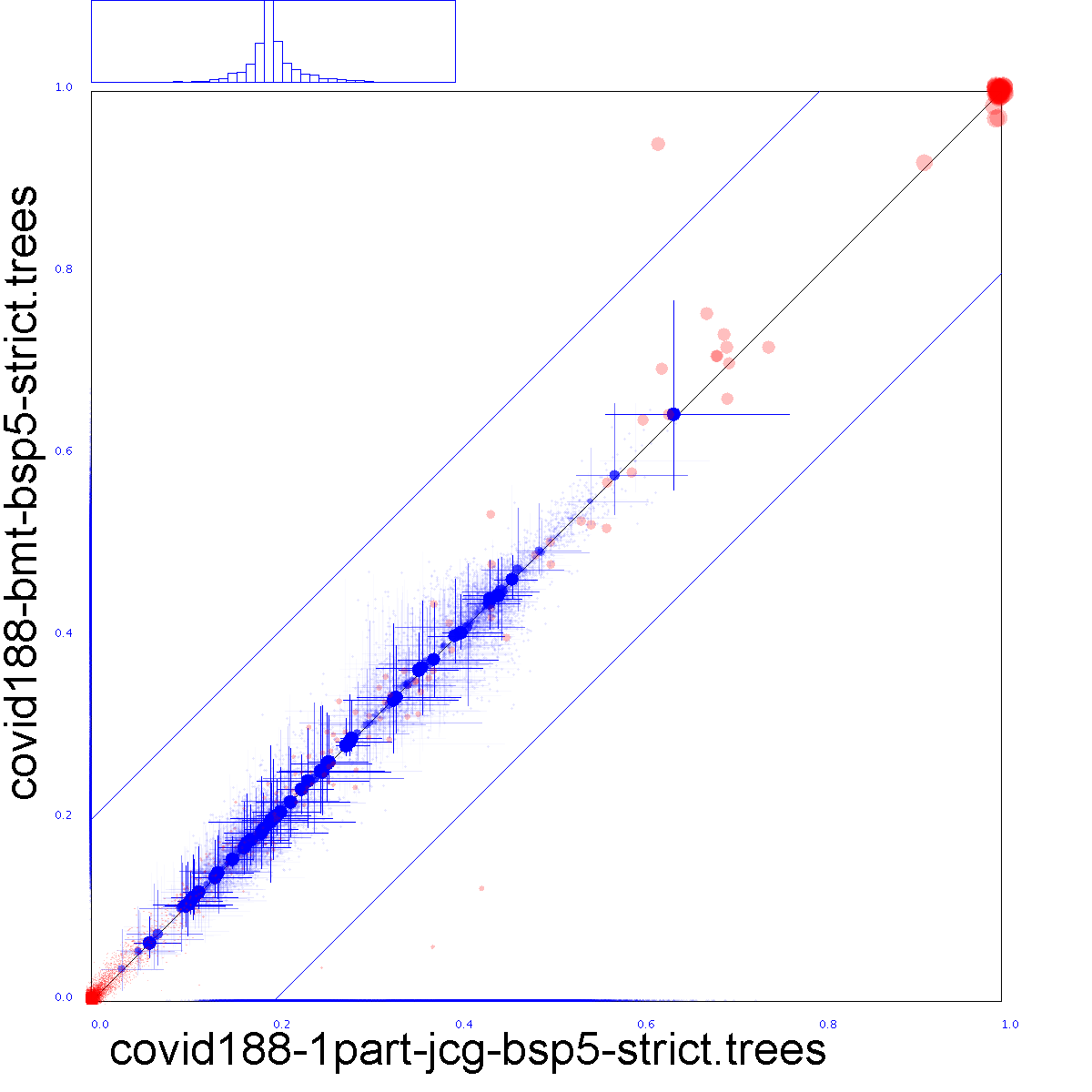 |
| x | 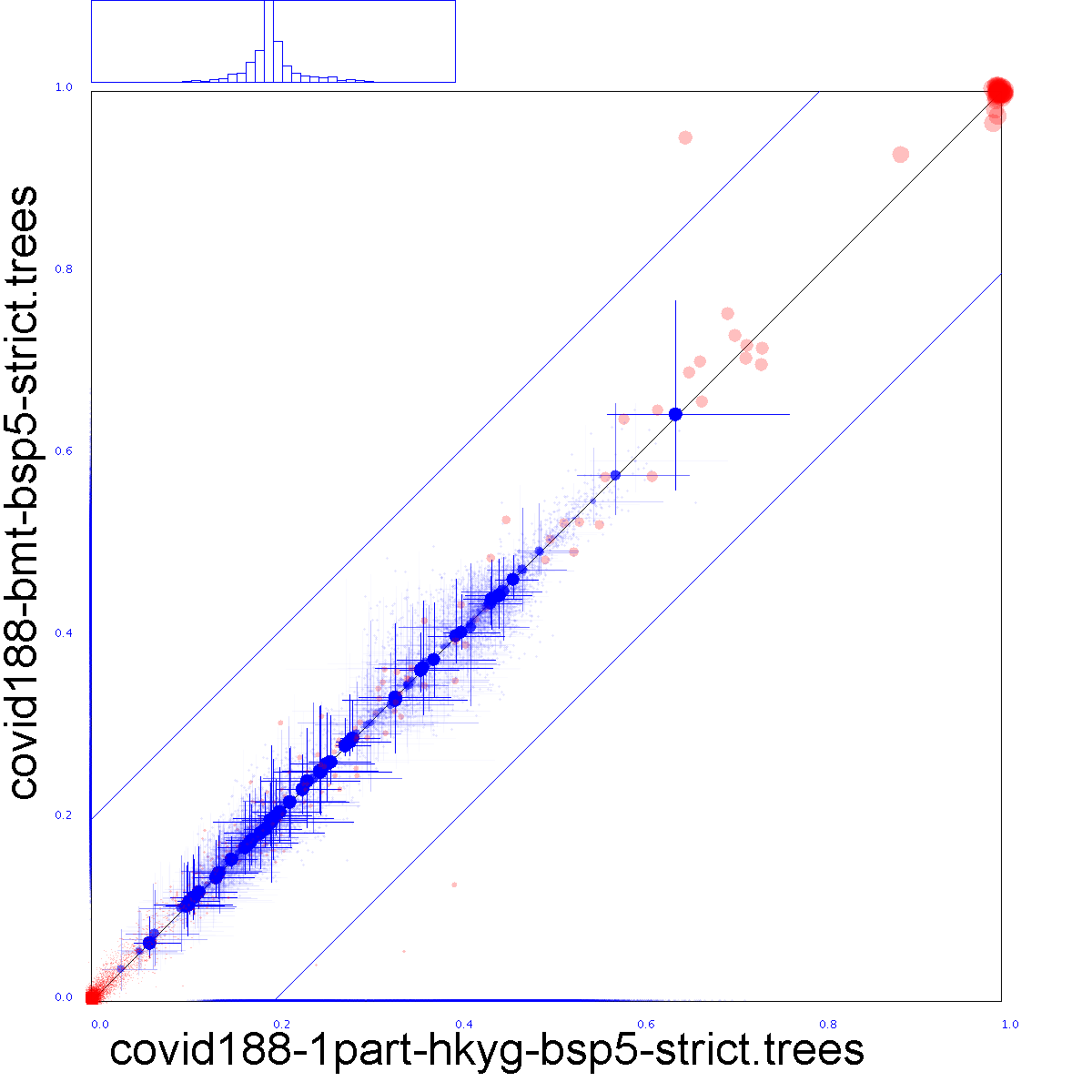 |
Screen output can look something like this:
Args:[-tree1, results3/2state.trees, -tree2, results2/2state.trees, -png, /tmp/diff.png, -out, /tmp/out.txt]
Processing results3/2state.trees
Processing 381 trees from file after ignoring first 10% = 42 trees.
Processing results2/2state.trees
Processing 354 trees from file after ignoring first 10% = 39 trees.
Writing to file /tmp/out.txt
Clade of interest (>25% difference): {Chongqing/IVDC-CQ-001/2020,Hangzhou/HZCDC0012/2020,USA/UPHL-01/2020} 0.4304461942257218 0.10734463276836158
Maximum difference in clade support: 0.7613995284487745
Done
Some things to note:
- Any clades with more than 25% difference in support between the tree sets will be listed. The two numbers at the end of the line represent clade support for set 1 and set 2. If there are clades with more than 25% support this suggests considerable difference in posterior tree sets.
- The text file (specified with
-out /tmp/out.txt) contains all clades and their supports. Since there can be quite a lot of clades, especially when there are many taxa, it is recommended to use the-outoption, otherwise that information is written on screen. - At the end of the run, the maximum number difference of clade support is printed. If that difference is more than 25% this suggests considerable difference in posterior tree sets.
