1 August 2019 by Remco Bouckaert
The effective sample size (ESS) is an important diagnostic for MCMC analyses to determine whether the run converged. There is already information on this page, but here we discuss a few more things you can consider to increase the ESS.
Increase BEAST performance
Often, you can increase speed of the chain by running it more efficiently, as outlined in the performance suggestions page. Essentially, you want to use the BEAGLE library, and use as many threads as help increase the number of MCMC samples per million states, which are reported on screen when BEAST runs.
Reduce complexity of the model.
In general, you want to make sure the analysis runs fine with the simplest model first before adding more complexity to it, so initially
- Use HKY instead of GTR.
- If you have many partitions, do not estimate mutation rates for partitions.
- Use strict clock instead of relaxed clock
- Use the simplest tree prior, i.e., Yule or Coalescent with constant population size. Using a birth/death model when there is little signal for the death rate will results in everything mixing, except for the prior and posterior.
Once the analysis converges with these settings, you can change aspects of the model to the point where the chain keeps mixing and giving good ESSs.
Check your model is valid
If your model is set up with unidentifiable parameters all of the above will not help – even if the model has identifiable parameters, in practice the signal in the data may be too weak to be able to estimate them
An example of actual unidentifiable parameters is when the substitution rates and clock rates are both estimated for all partitions (see this post for details on the different rates). This shows up in Tracer as low ESSs for the rate parameters, but reasonable ESSs for tree likelihoods.
An example of often hard to estimate parameters are the birth and death parameters of birth/death models. This shows up in Tracer as low ESSs for the prior and the posterior, but most other values mixing reasonably.
Balance operator weights
If only a few parameters (like the kappa and base frequencies parameters for the HKY substitution model) have low ESSs compared to other entries in the trace log, this can be fixed by increasing the operator weight. You can do this in BEAUti in the operators panel – by default, the panel is not visible, but by going to the View/Show Operators panel menu, you can make it appear.
Alternatively, you can directly edit the XML, go to the section with operators and change the weight:
<operator weight="1.0" ... >Tune operator parameters
At the end of a BEAST run, an operator report is printed to screen with some suggestions on how to change tuning parameters for each of the operators. It looks something like this:
Operator Tuning #accept #reject Pr(m) Pr(acc|m)
ScaleOperator(kappaScaler) 0.43252 244 250 0.01020 0.49393 Try setting scaleFactor to about 0.187
ScaleOperator(shapeScaler) 0.24783 387 184 0.01020 0.67776 Try setting scaleFactor to about 0.061
ScaleOperator(clockRateScaler) 0.63817 488 975 0.03061 0.33356
UpDownOperator(upDown) 0.70130 128 1383 0.03061 0.08471 Try setting scaleFactor to about 0.837
ScaleOperator(popSizesScaler) 0.24438 3974 3532 0.15306 0.52944 Try setting scaleFactor to about 0.06
DeltaExchangeOperator(groupSizesDelta) 6.16541 362 2177 0.06122 0.16628 Try setting delta to about 15.0
ScaleOperator(treeRootScaler) 0.69202 376 1167 0.03061 0.24368
Uniform(unknown) - 6678 8673 0.30612 0.43502
SubtreeSlide(unknown) 7.02560 2075 5624 0.15306 0.26952
Exchange(narrow) - 2323 5323 0.15306 0.30382
Exchange(wide) - 34 1545 0.03061 0.02153
WilsonBalding(unknown) - 24 1475 0.03061 0.01601
Tuning: The value of the operator's tuning parameter, or '-' if the operator can't be optimized.
#accept: The total number of times a proposal by this operator has been accepted.
#reject: The total number of times a proposal by this operator has been rejected.
Pr(m): The probability this operator is chosen in a step of the MCMC (i.e. the normalized weight).
Pr(acc|m): The acceptance probability (#accept as a fraction of the total proposals for this operator).
In this case, it suggests setting scaleFactors for various scale operators, and setting the delta for a delta exchange operator. Setting tuning parameters of operators helps operators to balance the boldness of the MCMC proposal and acceptance of such proposals.
Use operators that move multiple parameters
In Tracer, you can use the joint marginal panel to show correlation between the different parameters in your analysis. Here, we see how clock rate, tree heigh and population size are correlated:
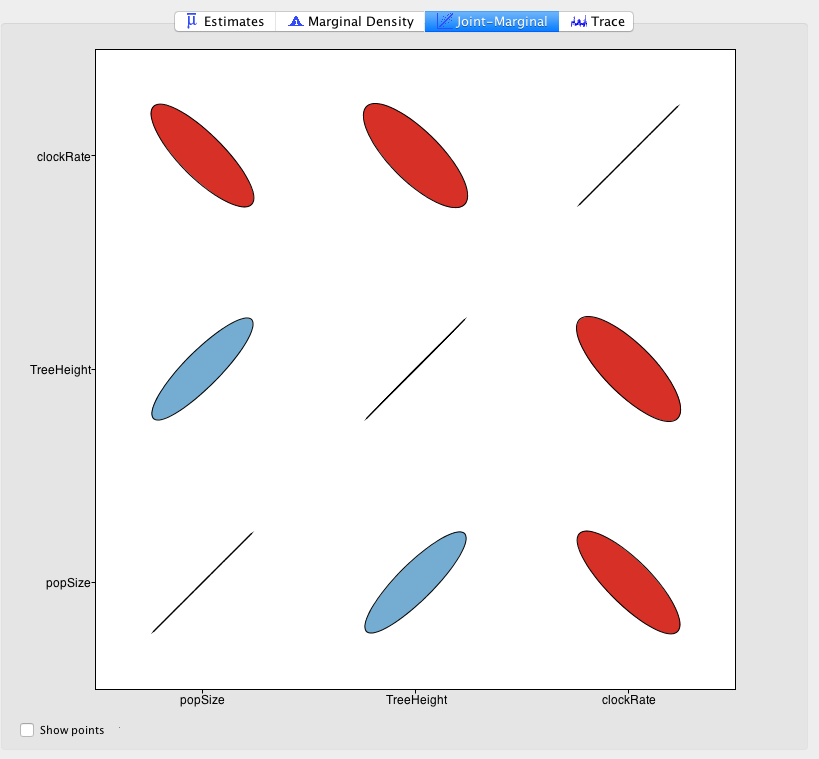
If the correlation is strong enough, and operators only work on one parameter at the time, it may prevent mixing. An operator that helps such mixing is the up-down operator, which is a scale operator that moves various parameters at the same time. You can add operators to the XML file, or add parameters to existing operators to help mixing.
<operator spec="beast.evolution.operators.UpDownOperator" scaleFactor="0.75" weight="3.0">
<up idref="Tree"/>
<up idref="popSize"/>
<down idref="clockRate"/>
</operator>For the up-down operator, you should group all parameters with positive correlation (positive slope of elipse on Tracer, here tree height and population size) in the up elements, and the negative correlated parameters (here the clock rate) in the down element. The Up/Down operator has the following attributes:
- scaleFactor (double): magnitude factor used for scaling (required)
- up (state node that can be scaled, like real parameter or tree): zero or more items to scale upwards (optional)
- down (state node): zero or more items to scale downwards (optional)
- optimise (boolean): flag to indicate that the scale factor is automatically changed in order to achieve a good acceptance rate (optional, default: true)
- elementWise (boolean): flag to indicate that the scaling is applied to a random index in multivariate parameters (optional, default: false)
- upper (Double): Upper limit of scale factor (optional, default: 1.0)
- lower (Double): Lower limit of scale factor (optional, default: 0.0)
- weight (Double): weight with which this operator is selected (required)
Use Coupled MCMC instead of MCMC
Another thing you might consider is using Coupled MCMC (also known as MCMCMC, MC3 and parallel tempering) instead of MCMC. It is a method that tends to help with badly mixing analyses and to some extend parallelyses the calculation. This allows for increasing the ESS (in real time) at the cost of extra computation and may be helpful if you can run it on a computer with many cores.
There is a tutorial for the Bayesian Skyline with Coupled MCMC: https://taming-the-beast.org/tutorials/CoupledMCMC-Tutorial/
