1 August 2024 by Remco Bouckaert
Usually, when running an MCMC analysis, the first thing to do is open Tracer to check the trace log. The main things to look out for are
- are the effective sample sizes (ESSs) high enough, especially for the posterior, prior and likelihood, and
- are independent runs converging to the same distribution for all parameters.
And if one of these two checks fails, usually the first thing to try is to resume the MCMC to obtain a longer chain. The thing that is missing from this process is a sense of how well the tree sample is mixed. The tree height, tree length and tree prior entries in the trace log only capture summary statistics of the sampled trees, but by themselves do not provide a good insight in how well the trees mix.
Automatic stopping
Recently, a new package became available that tries to address all of the above:
- it automatically assesses how long to run an MCMC before it can be stopped, and
- it considers metrics on tree space.
It achieves this by running two MCMC chains in parallel and periodically checks various stopping criteria. The default stopping criteria are:
TraceESS: this measures the ESS on selected traces, by default only posterior, likelihood and prior, but more can be added. There are exceptions where the ESS does not tell about the quality of mixing, such as population sizes for the Bayesian skyline plot, or indicator variables that are stuck at true or false in methods that us stochastic variable selection (e.g. bModelTest, DTA). Since it is not possible beforehand to determine whether a parameter should be included or not, only posterior, likelihood and prior are set up by default, since we know that the ESSs of those metrics should be high.TreePSRF: this tracks two metrics (1) a metric that compares the variance of RNNI distances within the individual chains with that of the variance when the tree sets of the individual chains are merged. This is similar to the R-hat metric (Gelman & Rubin, 1992). (2) tree ESS (as per Lanfear et al, 2016) but with growing number of focal trees for stability of ESS estimates.
When all stopping criteria pass, the MCMC stops and produces a combined trace and tree logs with burn-in removed from the two independent MCMC chains. So, there should be no more need to resume an MCMC analysis when using the ASM method.
For more details, see Berling et al, 2024.
In practice
Instructions for usage for the ASM package is available [here](](https://github.com/rbouckaert/asm/blob/main/README.md). Essentially, it is a matter of installing the package and selecting ASM on the MCMC panel in BEAUti.
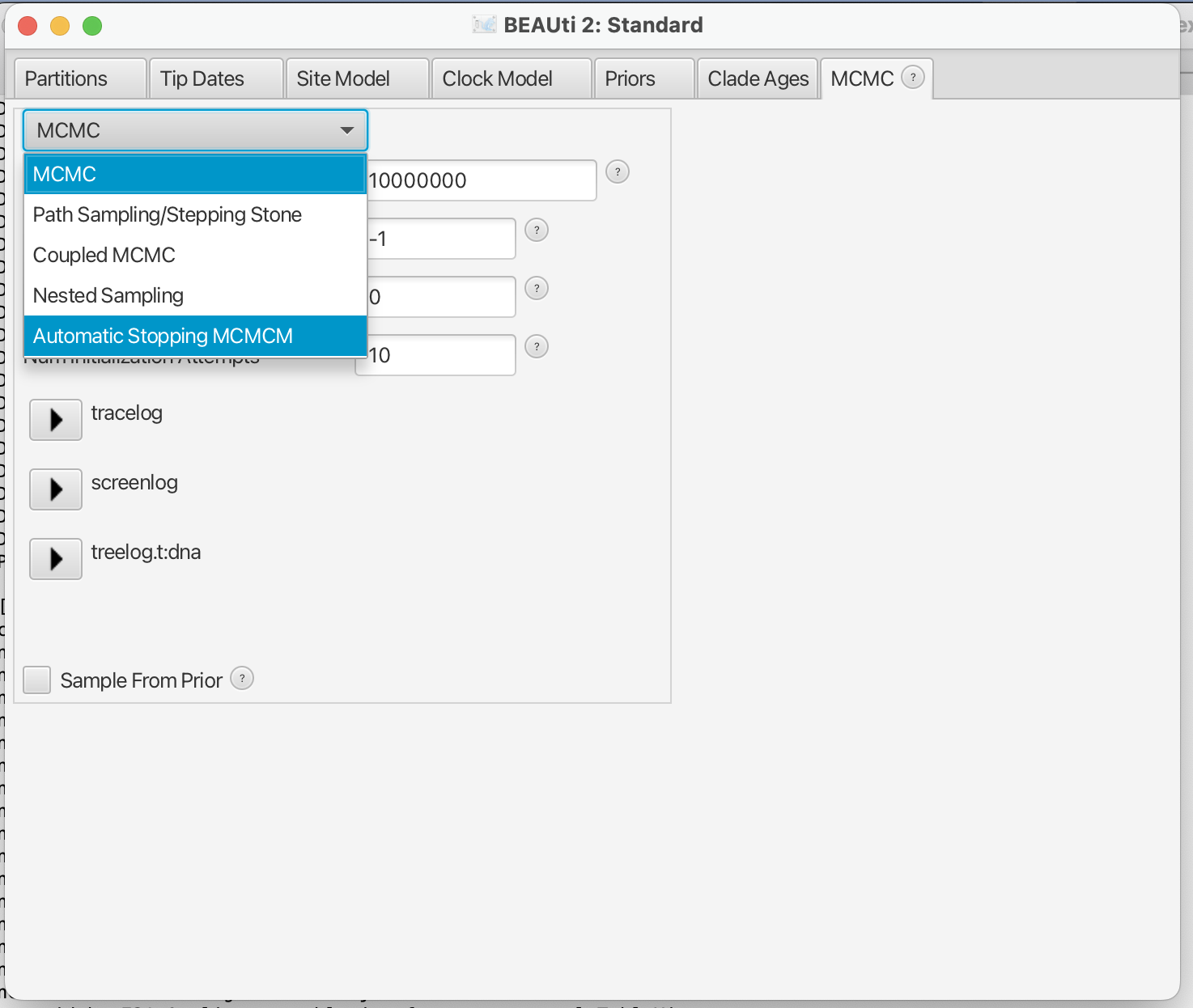
You can set parameters by clicking the TreePSRF button for the Gelman-Rubin-like criterion or TraceESS button for the trace-ESS criterion.
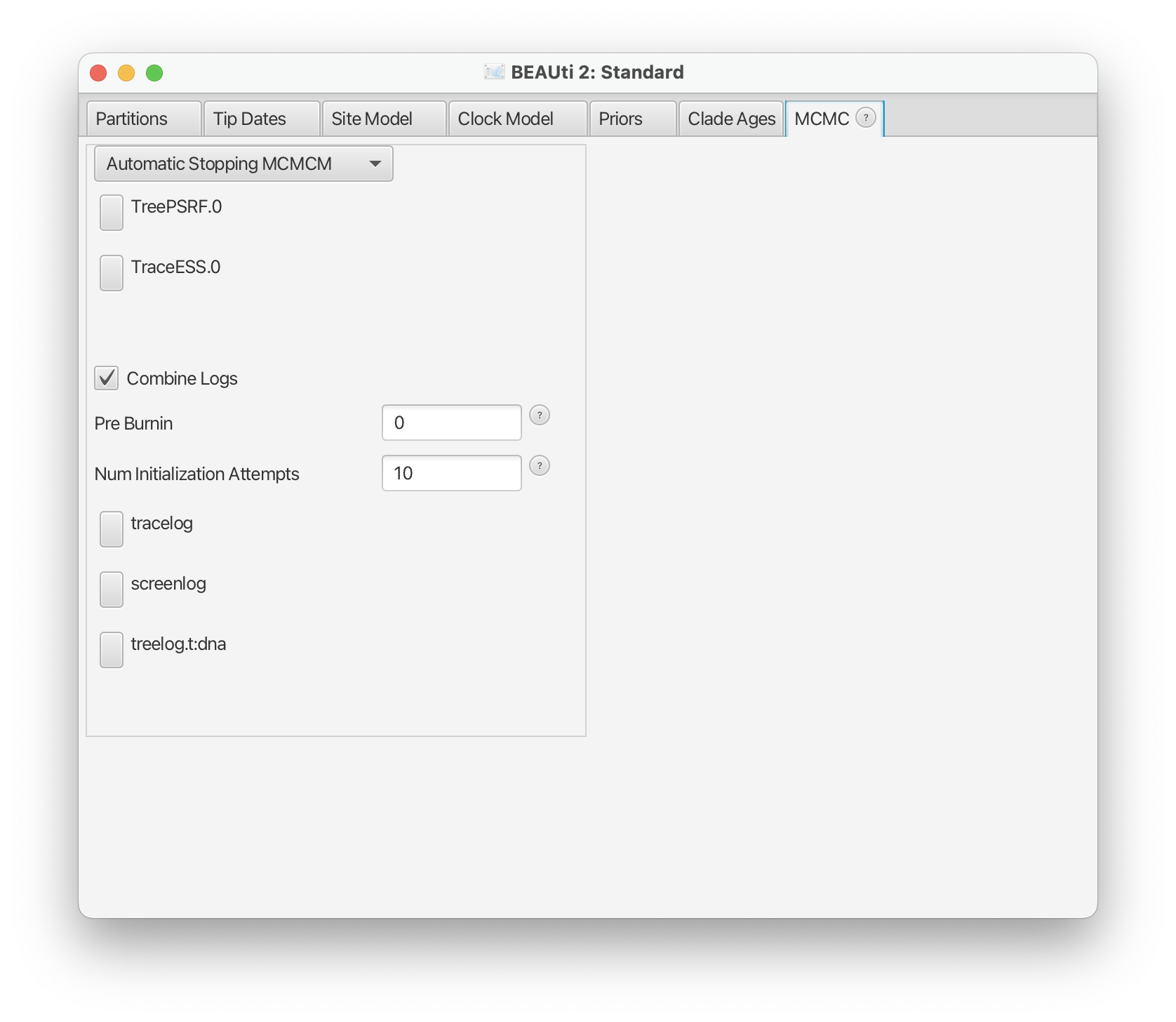
Interpreting screen output
Screen output may look something like this:
Check 19 burnin:[14, 14]psrf1mean = 1.472 reset start -1 10.0 5.1 10.0 :5.1 in 5 mseconds
Check 20 burnin:[15, 15]psrf1mean = 1.399 reset start -1 7.2 5.3 10.0 :5.3 in 5 mseconds
Check 21 burnin:[15, 15]psrf1mean = 1.426 reset start -1 10.9 7.2 12.0 :7.2 in 5 mseconds
Check 22 burnin:[16, 16]psrf1mean = 1.466 reset start -1 12.0 12.0 12.0 :12.0 in 3 mseconds
Check 23 burnin:[17, 17]psrf1mean = 1.525 reset start -1 8.8 12.0 8.9 :8.8 in 7 mseconds
Check 24 burnin:[18, 18]psrf1mean = 1.504 reset start -1 7.6 12.0 6.9 :6.9 in 18 mseconds
Let’s have a look at the first entry:
Check 19 burnin:[14, 14]psrf1mean = 1.472 reset start -1 10.0 5.1 10.0 :5.1 in 5 mseconds
Check 19 check number
burnin:[14, 14] log items removed as burnin, first number for chain1, second for chain2
psrf1mean = 1.472 potential scale reduction factor — this statistic represents a Gelman-Rubin like statistic for the trees. It should go towards 1 at convergence.
reset start -1 whether the start of the psrf is recalculated from the start
10.0 5.1 10.0 ESSs of the statistics specified in the asm.inference.TraceESS statistic
:5.1 minimum of the ESSs of the asm.inference.TraceESS statistic. This should go to the threshold specified in the targetESS attribute.
in 5 mseconds time used to calculate the statistics
Sampling from prior
Normally, to get a sample from the prior for an MCMC run, the -sampleFromPrior flag can be used for BEAST.
This fails with ASM, so to obtain a sample from the prior, the XML must be edited to produce a sample from the prior, like so:
- move the element with
id="likelihood"including all of its sub-elements to outside therunelement, say at the end of the file, just before the</beast>line. - remove
likelihoodfrom thetracesattribute of theTraceESSstopping criterion.
If you get this kind of error:
Check 0 java.lang.IllegalArgumentException: Could not find label likelihood in trace log. Use one of [Sample, posterior, prior, ...
the traces attribute is not specified, and the default (i.e. posterior,likelihood,prior) will be used. To fix this the traces="posterior,prior" must be added to the TraceESS stopping criterion, so it looks something like
<stoppingCriterion spec="asm.inference.TraceESS" targetESS="100" traces="posterior,prior"/>
References
Lars Berling, Remco Bouckaert, and Alex Gavryushkin. An automated convergence diagnostic for phylogenetic MCMC analyses IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2024 Sep 10. DOI (BioRxiv preprint)
Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Statistical science. 1992 Nov;7(4):457-72.
Lanfear R, Hua X, Warren DL. Estimating the effective sample size of tree topologies from Bayesian phylogenetic analyses. Genome biology and evolution. 2016 Aug 1;8(8):2319-32.
